Today, we're excited to announce the open-source release of two critical resources for Indian healthcare digitisation: Parrotlet-e, a state-of-the-art Indic medical embedding model, and Eka-IndicMTEB, a sizable Indic medical terms embedding benchmark dataset. Try out the model on HuggingFace.
These releases address a fundamental challenge in India: the ability to understand and process medical information across multiple languages and scripts while maintaining semantic accuracy.
Parrotlet-e model is openly available on AIKosh and HuggingFace
Eka-IndicMTEB evaluation dataset is openly available on AIKosh and HuggingFace
“Patient ka sir ghoom raha hai, BP low hai, sugar thoda badha hua hai, next week recheck karenge.”
That’s how a real consultation sounds in a busy clinic in North India — a fluid mix of Hindi or any local language and English, clinical shorthand, and regional phrasing. India's healthcare system operates in a uniquely complex linguistic environment. Medical scribe systems that aim to convert doctor-patient conversations into structured records must navigate this linguistic diversity before they can extract meaningful information.
The core problem isn't just translation—it's semantic understanding. When a patient describes their condition as "मधुमेह" (Hindi), "diabetes" (English), or "ಸಕ್ಕರೆ ಕೈಲೆ" (Kannada), healthcare systems need to recognise these as identical concepts. This semantic alignment is essential for:
Without robust multilingual medical entity understanding, each of these systems operate in isolation, creating data silos that fragment India's healthcare infrastructure.
Parrotlet-e is designed to solve this exact problem. It's an embedding model that represents medical entities in a shared semantic space, where similar concepts cluster together regardless of the language or script used to express them.
Comprehensive Indic Coverage: Unlike existing medical embedding models optimised primarily for English, Parrotlet-e handles entity-level representations across Indian languages—both in native scripts and romanised forms. It currently handles 12 Indic languages, namely: Hindi, Marathi, Malayalam, Kannada, Tamil, Telugu, Gujarati, Punjabi, Odia, Assamese, Bengali, and Urdu.
Real-world Robustness: The model accounts for the messy reality of healthcare documentation: abbreviations, spelling variations and colloquial variations.
Clinical Focus: Built specifically for medical entities such as symptoms, diagnoses and anatomical structures, with alignment to SNOMED CT [1] terminology.
Creating training data for a multilingual medical embedding model required a multi-faceted approach:
Foundation from Established Terminologies: We started with SNOMED CT official terminology and UMLS [2] as our base, ensuring alignment with international standards.
Clinical Abbreviations: Healthcare professionals very frequently use abbreviations for medical concepts. We systematically collated common abbreviations used in Indian clinical practice.
Expert Medical Annotations: Our in-house medical team contributed multilingual term annotations, ensuring clinical accuracy across languages.
Proprietary Clinical Data: EkaCare's extensive databases provided real-world term variations observed in actual healthcare settings.
Parrotlet-e is fine-tuned from bge-m3 [3] using weakly supervised contrastive learning with Multi-Similarity Loss [4] and a batch size of 1024. Contrastive learning trains the model to map similar terms closer in its semantic representation space while pushing dissimilar terms farther apart. Generally, contrastive learning approaches treat every similar (positive) and dissimilar (negative) example similarly. Therefore, we use Multi-Similarity Loss which weighs example pairs by their relative difficulty rather than treating all positives and negatives uniformly. It automatically emphasizes harder cases: difficult positives that require finer discrimination and challenging negatives that could be mistaken for matches. This means the model focuses more attention on challenging cases—difficult positives requiring fine-grained discrimination and hard negatives that could easily be confused with true matches. The result is a more robust embedding space that handles the subtle distinctions critical in medical terminology. Overall, for training, we collated a dataset comprising 18 million positive pairs across languages and scripts.
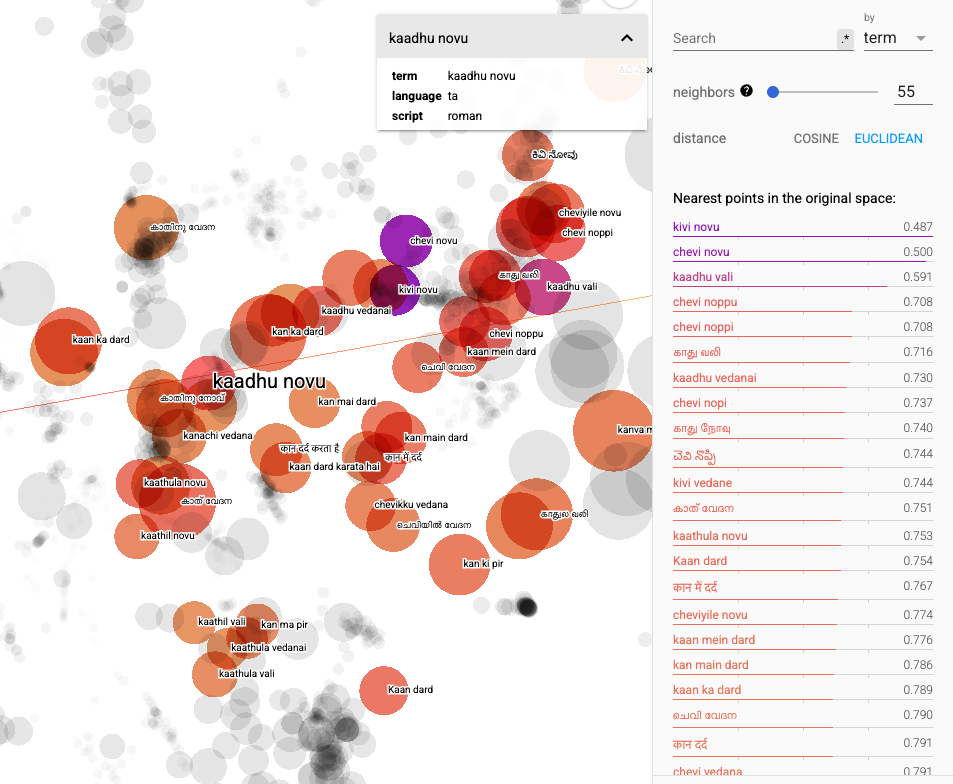
An example of T-SNE plot of our dataset is shown below. We can clearly see how terms in different languages and scripts referring to a medical concept (ear pain in this case) groups together.
Releasing a model is only half the story. To drive progress in multilingual medical AI, the community needs a rigorous way to evaluate performance. That's why we're also introducing Eka-IndicMTEB—EkaCare's Indian Multilingual Terms Embedding Benchmark.
Eka-IndicMTEB contains 2532 carefully curated query entities spanning multiple Indian languages and scripts. Each entity has been tagged with its corresponding SNOMED CT identifier by a medical professional, ensuring clinical accuracy.
The benchmark is designed to reflect how medical language truly appears in Indian healthcare:
Existing medical embedding benchmarks are predominantly English-focused and don't capture the linguistic complexity of Indian healthcare. Eka-IndicMTEB fills this gap by providing:
We hope Eka-IndicMTEB becomes a foundation for advancing multilingual medical entity embedding in India.
The dataset is publicly available on AIKosh and HuggingFace. The dataset contains three subsets
We conducted comprehensive evaluations of Parrotlet-e against several strong baselines: SapBERT [5] (the current state-of-the-art for English medical entity embeddings), EmbeddingGemma [6] (Google's embedding model), IndicBERTv2 [7] (a model pre-trained on Indic languages), and the base bge-m3 model. All models were evaluated using KARMA on Eka-IndicMTEB, with metrics computed at Recall@1, Recall@3, and Recall@5—representing the percentage of queries where the correct SNOMED CT entity appears in the top 1, 3, and 5 retrieved results, respectively.
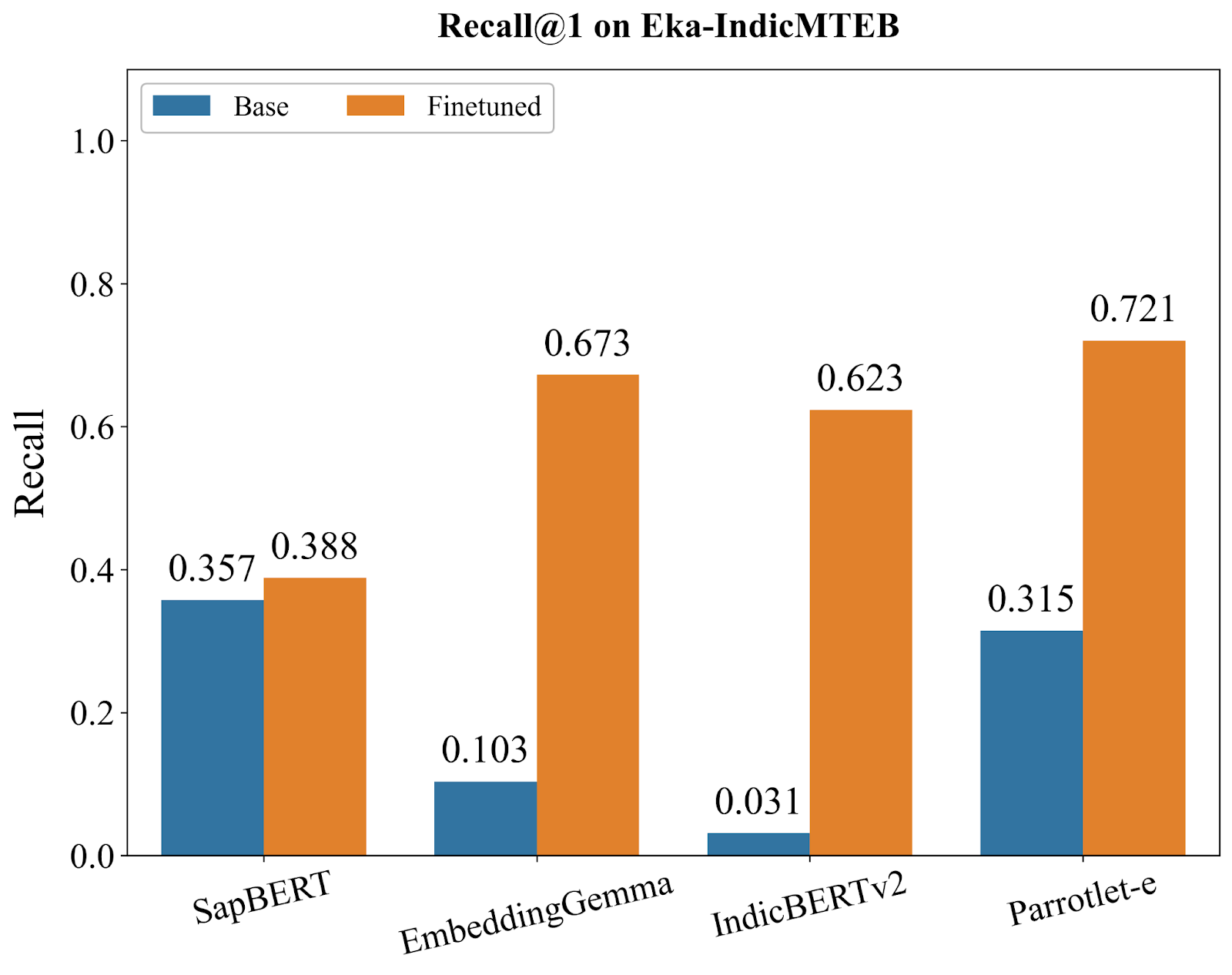
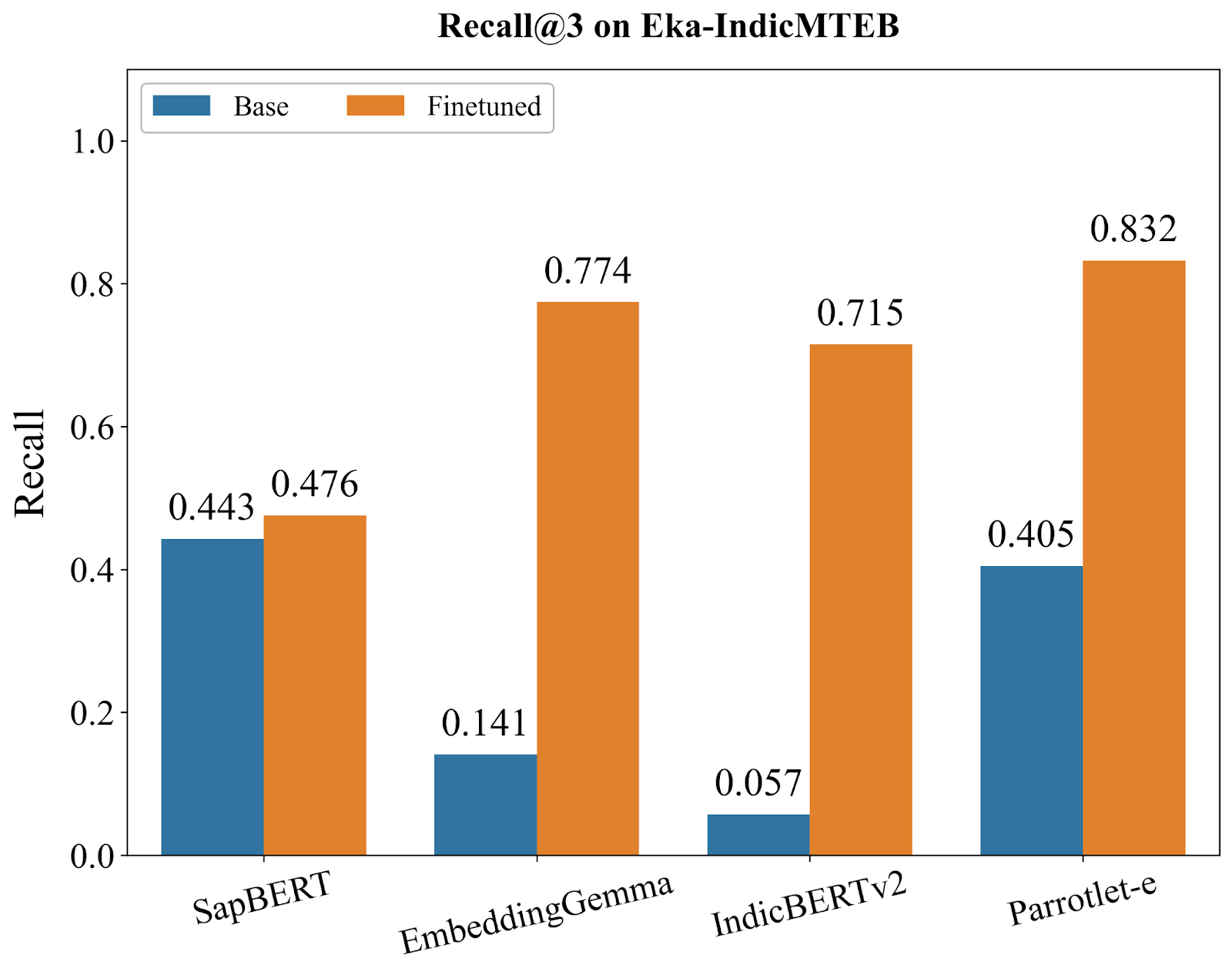
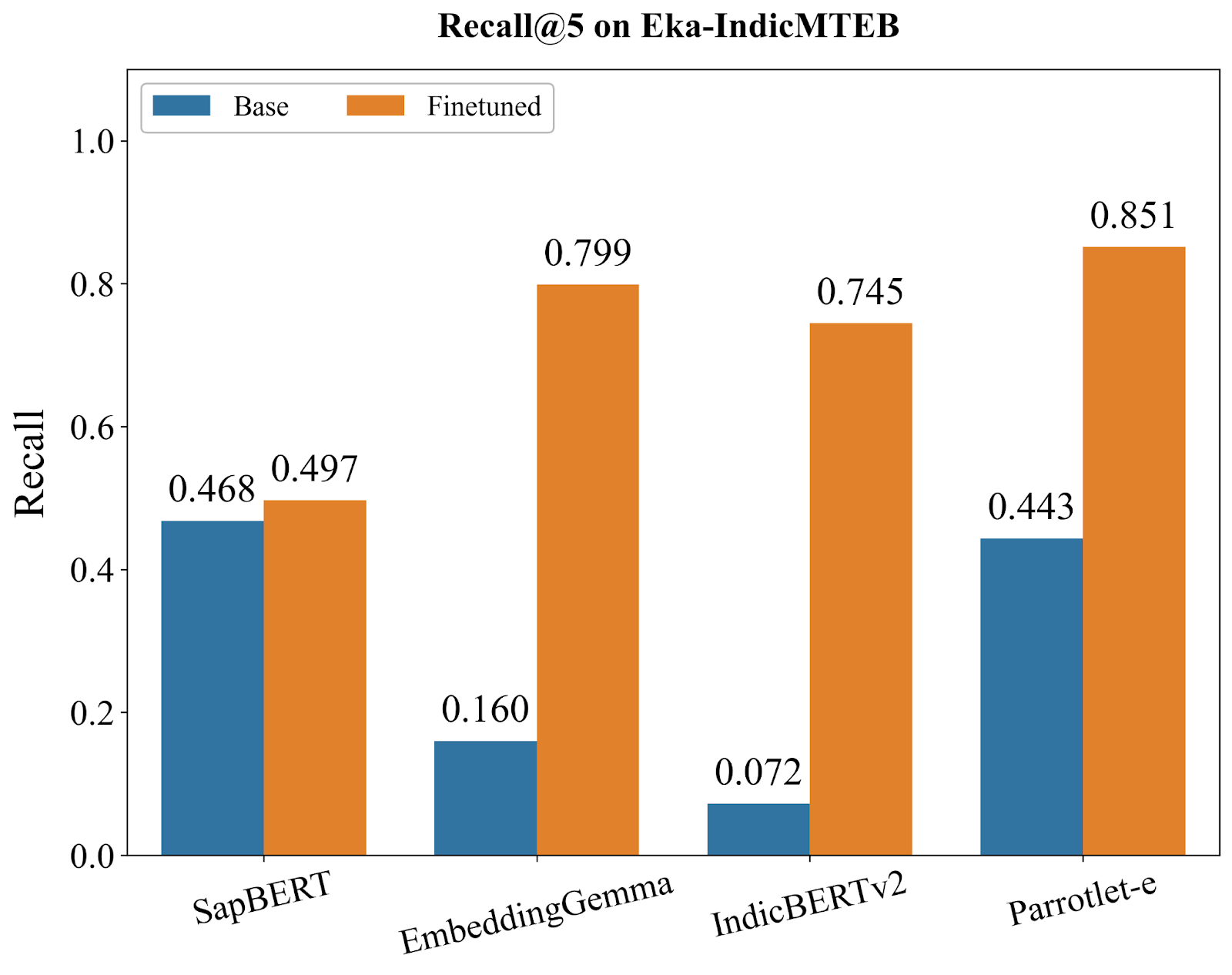
The benchmark results reveal three distinct model archetypes, each telling us something important about what's needed for multilingual medical entity understanding:
SapBERT, despite being the gold standard for English medical embeddings, shows the smallest improvement from finetuning (+8.7% at R@1). This isn't a failure of our training approach—it's a fundamental limitation. SapBERT's architecture and pre-training were optimized exclusively for English medical terminology. When confronted with Hindi, Tamil, Kannada, or even romanised Indic terms, it lacks the foundational linguistic representations needed to benefit from additional training.
The plateau in SapBERT's performance demonstrates that clinical domain expertise alone is insufficient for multilingual medical AI. A model can deeply understand "diabetes" while completely missing "मधुमेह" or "diabeetus" (a common phonetic misspelling). This insight validates our core thesis: India's healthcare digitisation requires models built from the ground up for linguistic diversity.
EmbeddingGemma and IndicBERTv2 represent the opposite starting point—strong multilingual capabilities with virtually no clinical knowledge. Their base models achieve just 10.3% and 3.1% R@1, respectively. Yet after finetuning, they surge to 66.2% and 62.3%, demonstrating massive improvements.
This dramatic transformation reveals a critical insight: models with robust Indic language understanding can rapidly acquire clinical domain knowledge through targeted training. These models already know how to represent "मधुमेह", "डायबिटीज", and "sugar problem" in a shared semantic space—they just need to learn which concepts are medically equivalent.
The massive gains indicate that the linguistic understanding was there all along; these models simply lacked the clinical reasoning to connect symptoms, diagnoses, and anatomical structures. Our training data provided that missing clinical layer, unlocking their potential for medical entity matching.
Parrotlet-e (built on bge-m3) occupies a unique position: its base model already achieves 31.5% R@1, substantially higher than EmbeddingGemma (10.3%) or IndicBERTv2 (3.1%), though lower than SapBERT (35.7%). This indicates bge-m3 starts with both reasonable clinical understanding AND multilingual capability—a rare combination that provides the ideal foundation for our task.
After finetuning, Parrotlet-e reaches 72.1% R@1, decisively outperforming all alternatives. More importantly, it sustains this advantage across all recall levels, achieving 83.2% R@3 and 85.1% R@5. These numbers matter in practice: in a medical coding interface, users almost always examine the top 3-5 results. An 85% chance of seeing the correct entity in that initial view means the system is genuinely useful, not just academically interesting.
To understand where each model truly excels and where critical gaps remain, we conducted granular error analysis by breaking down Recall@1 performance across three distinct query categories:
Note that English only queries also include abbreviations.
These results reveal not just which model is "best," but when each model succeeds or fails—critical insights for deploying these systems in production.
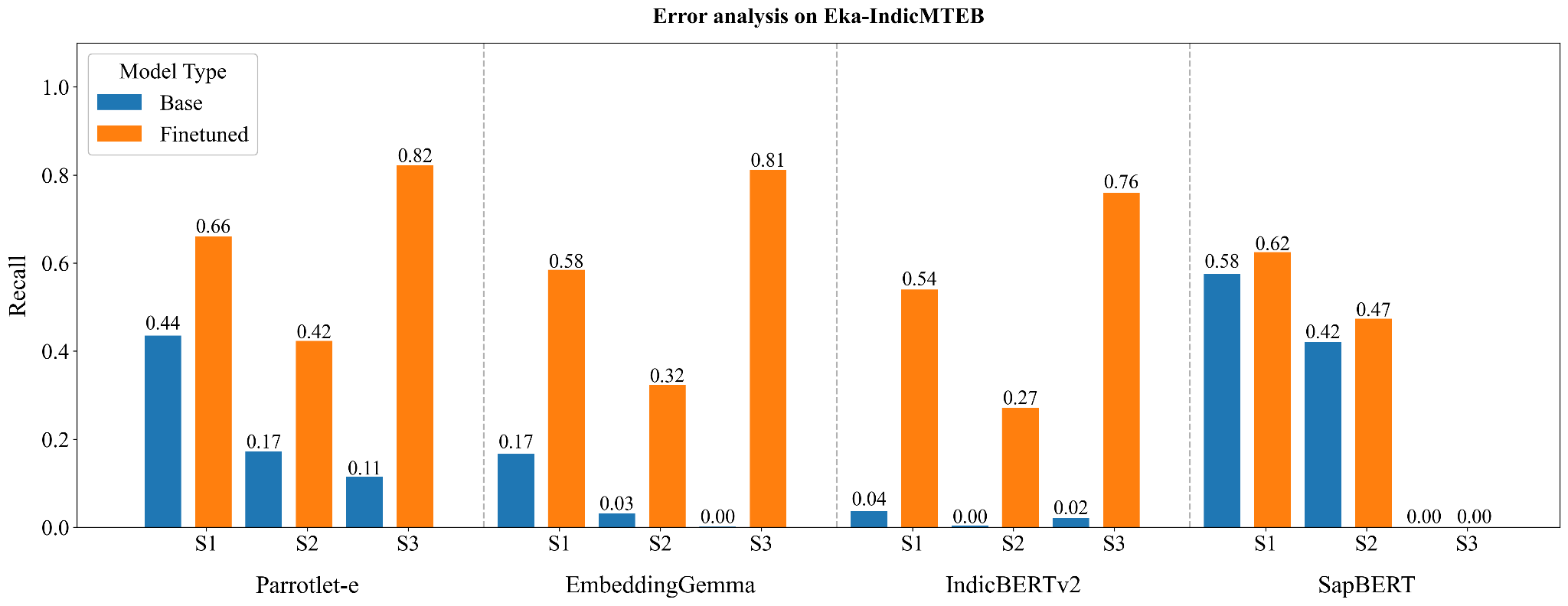
English Medical Terms Subset:
Clinical Abbreviations (Hardest Category):
Indian Languages:
We examine deeply why the best performance on Indic languages is higher than that on English only subset. The primary reason for this observation is the presence of abbreviations with the English terms, on which the performance is the worst.
Key Insights from error analysis:
Top-K Accuracy Matters More Than R@1: The progression from 72.1% (R@1) to 83.2% (R@3) to 85.1% (R@5) suggests that Parrotlet-e's ranking is strong—the correct entity is almost always in the top few results, even if not always first. For medical coding interfaces where users review multiple suggestions, this ranking quality is crucial.
The Gap Between Base and Finetuned Reveals Training Quality: Models that improve dramatically (IndicBERTv2: +1909%) had the linguistic foundation but zero clinical knowledge. Models that barely improve (SapBERT: +8.7%) hit an architectural ceiling. Parrotlet-e's balanced improvement (+129%) suggests optimal knowledge transfer without overfitting.
Consistent Lead Across Metrics: Parrotlet-e doesn't just win at R@1—it maintains superiority at R@3 (83.2% vs. 75.7% for second-best) and R@5 (85.1% vs. 78.5% for second-best). This consistency indicates the model isn't just memorizing specific entity pairs but learning generalizable medical-linguistic patterns.
Real-World Readiness: An 85.1% R@5 score means that in a production medical coding system, 85 out of 100 clinical terms—across multiple languages, with spelling variations, abbreviations, and colloquialisms—will surface the correct SNOMED CT code in the top 5 suggestions. This crosses the threshold from "research prototype" to "production-ready tool."
Parrotlet-e enables several critical applications in Indian healthcare:
Medical coding is the backbone of digital health records, insurance processing, and clinical decision support. With Parrotlet-e, you can directly search for the nearest SNOMED CT entity using a query in any supported language:
# Query in Hindi
query = "मधुमेह की जांच"
# Returns closest SNOMED CT concepts:
# 1. Diabetes mellitus screening (SNOMED: 268547008)
# 2. Blood glucose measurement (SNOMED: 33747003)
# 3. Hemoglobin A1c measurement (SNOMED: 43396009)This enables automatic medical coding systems that work across languages, removing a major bottleneck in healthcare digitization.
AI-powered medical scribes need to understand clinical concepts before they can structure conversations into medical records. Parrotlet-e provides the semantic foundation for:
State-of-the-art language models often misinterpret colloquial medical terms, treating them verbatim rather than understanding their semantic meaning. Parrotlet-e enables more sophisticated Retrieval-Augmented Generation (RAG) pipelines:
Healthcare digitization in India requires tools that understand India's linguistic reality. By open-sourcing Parrotlet-e and Eka-IndicMTEB, we're providing the developer community with:
Model: Parrotlet-e is available on AIKosh and HuggingFace
Benchmark: Eka-IndicMTEB dataset is available at the following links at AIKosh and HuggingFace
We thank IndiaAI Mission for supporting this work by offering GPU compute subsidy. Their support enabled us to train Parrotlet-e across multiple Indic languages and conduct comprehensive evaluations on Eka-IndicMTEB. Many thanks to the AIKosh team as well for extending their support.
[1] https://www.nlm.nih.gov/healthit/snomedct/index.html
[2] https://www.nlm.nih.gov/research/umls/index.html
[3] Chen J. et al. (2024). M3-Embedding: Multi-Linguality, Multi-Functionality, Multi-Granularity — Text Embeddings Through Self-Knowledge Distillation.
[4] Wang, Z. et al. (2019). Multi-Similarity Loss with General Pair Weighting for Deep Metric Learning.
[5] Liu, H. et al. (2020). SapBERT: Self-alignment Pre-training for Biomedical Entity Representations.
[6] https://huggingface.co/google/embeddinggemma-300m
[7] https://huggingface.co/ai4bharat/IndicBERTv2-MLM-only
Parrotlet-e and Eka-IndicMTEB are released under cc-by-sa-4.0. We encourage responsible use and welcome contributions to improve these resources for the broader healthcare community.