Healthcare faces a significant obstacle in achieving meaningful digitisation (blog) and enabling structured information exchange using standardised medical terminologies (blog). AI systems capable of processing unstructured medical data offer a promising solution to these challenges. EkaCare remains dedicated to driving digital transformation across India's healthcare landscape. In our previous work, we have shown that purpose-driven small LLMs fine-tuned on domain-specific data can outperform state-of-the-art systems in healthcare applications (blog). We are now releasing an open-weight lite version of our vision LLM, designed to extract structured data from laboratory reports and digital prescriptions.
Download model (HuggingFace | AIKosh)
Parrotlet-v-lite-4b builds upon the MedGemma3 4B foundation architecture. We performed supervised fine-tuning (SFT) of the model using an extensive dataset of medical records that were carefully annotated by healthcare professionals. Prior to training, all personally identifiable information (PII) was systematically removed through a comprehensive redaction process.
The training process employed Low-Rank Adaptation (LoRA) parameter-efficient fine-tuning with a rank configuration of 128 and an alpha value of 256, while keeping all other model layers frozen. Training utilized a learning rate of 1e-5 with bf16 precision. Following training completion, the LoRA adapter layers were integrated with the base model for deployment and evaluation.
EkaCare medical report parsing dataset encompasses a wide range of medical documents, including laboratory reports, prescriptions, discharge summaries, and medical imaging scans. A portion of this dataset was reserved for model evaluation purposes (referred to as eval-dataset-internal). Furthermore, a curated subset of eval-dataset-internal is being made publicly available to enable community benchmarking and comparison studies (referred to as eval-dataset-public). The curation was such that eval-dataset-public comprises more diversity in formats, naturally leading to more complicated cases being selected.
A few samples from our publicly released dataset are shown below to highlight the complexity and diversity of the training material.
In order to evaluate the model's performance, we used eval-dataset-public. We have carefully separated this evaluation set from our training dataset to avoid any data contamination.
We implemented a rubric-based evaluation framework utilising large language models as automated judges to assess extraction performance. The framework assigns binary scores (true/false) based on whether specific information elements are correctly present in the model's parsed output. This approach addresses the challenge of evaluating extraction accuracy across heterogeneous medical documents, where traditional string-matching or alignment metrics are insufficient.
The evaluation process works as follows: each rubric contains multiple criteria that assess different aspects of information extraction. An LLM judge evaluates the model's output against each criterion, determining whether the required information is accurately extracted. The final score for each document is calculated as the proportion of criteria successfully met. In the current release, we focus exclusively on evaluating the correct identification and presence of information elements, without penalising for extraneous content. This methodology provides a scalable and consistent approach to measure extraction quality across diverse document formats including lab reports, prescriptions, and discharge summaries.
The results below are obtained using GPT-4o as the judge. All the evaluations are done using KARMA evaluation toolkit.
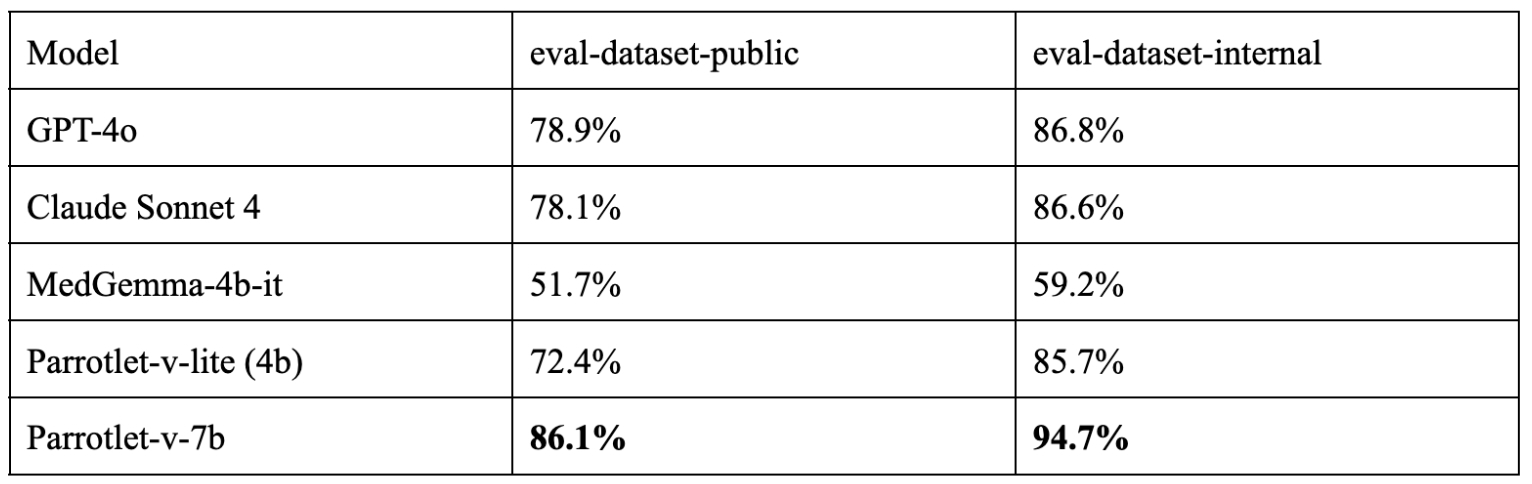
We observe that the accuracy of all the models on eval-dataset-public is reduced by nearly 10% compared to their accuracy on eval-dataset-internal. This is expected as mentioned earlier, that eval-dataset-public comprises more complex medical documents. However, the loss in accuracy is more prominent for the smaller model, Parrotlet-v-lite.
The best performing model is Parrotlet-v, which is EkaCare’s flagship model for medical report parsing accessible through an easy-to-use API.
Key Insights from Our Experimental Analysis: